
MultiCens
Multilayer network centrality measures to uncover molecular mediators of tissue-tissue communication.

Multilayer network centrality measures to uncover molecular mediators of tissue-tissue communication.
MultiCens is a versatile platform designed for the prediction of centrality scores associated with genes in various contexts, including specific tissues, regions, or layers, based on gene expression matrices. The platform offers three primary modes of centrality analysis:
MultiCens serves as a powerful tool for exploring the centrality of genes within complex multilayer networks, offering insights into their local, global, and query-set centrality characteristics based on gene expression data.
MultiCens offers a wide range of practical applications in diverse scientific domains. Some key applications include:
- Predicting hormone-responsive genes in a target tissue by providing inputs such as source and target tissue names, the mediator hormone, and a list of genes from both source and target tissues.
- Ranking hormone-responsive genes in the target tissue based on their responsiveness to the selected hormone.
- Identifying ligand-receptor relationships across multiple tissues, facilitating the study of molecular interactions.
- Shedding light on the complex process of inter-organ communication during metastasis, contributing to a better understanding of this intricate biological phenomenon.
- Investigating the reshaping of gene-gene networks between brain regions, particularly in the context of Alzheimer's disease.
- Applicability to other brain diseases involving multiple regions, thereby aiding in the study and potential treatment of such disorders.
- Extending the tool's utility to the analysis of multi-omics data, where each omics dataset is treated as a distinct layer in the analysis.
- This enables a comprehensive exploration of complex biological systems, offering insights into multi-dimensional molecular interactions.
Methods/Website/Software Development Supported Generously by: DBT/Wellcome Trust India Alliance Grant IA/I/17/2/503323.
"All users, including commercial users, can freely access this website."
| Select | Tissue/Region Name | Dataset | File | Remove |
|---|
| Tissue/Region Name | Centrality | DataSet | Result |
|---|---|---|---|
| Adrenal | Local | GTEx | Adrenal |
| Heart | Local | GTEx | Heart |
| Kidney | Local | GTEx | Kidney |
| Liver | Local | GTEx | Liver |
| Muscle | Local | GTEx | Muscle |
| Pancreas | Local | GTEx | Pancreas |
| Muscle_Pancreas | Local | GTEx | Muscle_Pancreas |
| Adipose_Adrenal | Global | GTEx | Adipose_Adrenal |
| Heart_Liver | Global | GTEx | Heart_Liver |
| Kidney_Liver | Global | GTEx | Kidney_Liver |
| Muscle_Stomach | Global | GTEx | Muscle_Stomach |
| Heart_Liver_Kidney | Global | GTEx | Heart_Liver_Kidney |
| Hypothalamus_Kidney_Liver | Global | GTEx | Hypothalamus_Kidney_Liver |
| Kidney_Heart_Muscle | Global | GTEx | Kidney_Heart_Muscle |
| Adrenal_Liver_Muscle_Pancreas | Global | GTEx | Adrenal_Liver_Muscle_Pancreas |
| Heart_Hypothalamus_Liver_Muscle | Global | GTEx | Heart_Hypothalamus_Liver_Muscle |
| Muscle_Pancreas | Query-Set | GTEx | Muscle_Pancreas |
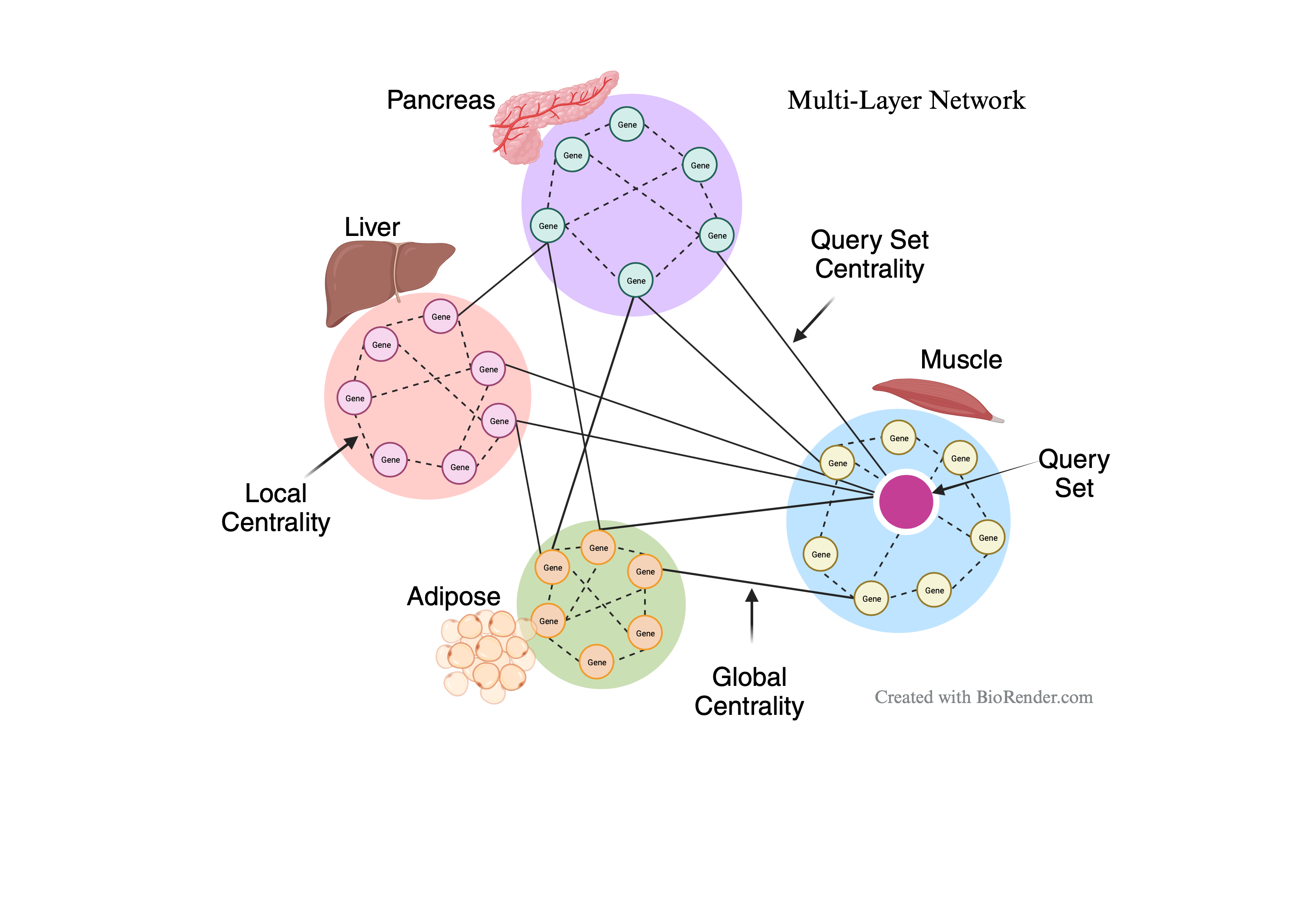
Interactions among biomolecules such as genes are crucial to a biological system, and a network of such interactions among genes residing in different tissues or regions within the body is called a multilayer network. Given that proper communication among different organs and tissues is essential for the healthy functioning of our body, and complex diseases often affect multiple organs or tissues, there is growing interest in constructing network models of genes residing in different tissues from multi-tissue genomic data. However, a major challenge lies in analyzing and extracting biological insights from such multi-tissue or multilayer network models. MultiCens offers a way to quantify the importance (also known as centrality) of genes (also known as nodes) within a multilayer network. MultiCens stands out as a robust tool designed to delve into the centrality of genes within intricate multilayer networks, particularly focusing on inter-tissue communication. It provides valuable insights into the local, global, and query-set centrality characteristics of genes, leveraging gene expression data for comprehensive analysis. With its capabilities, MultiCens empowers users to gain a deeper understanding of gene centrality dynamics across diverse biological contexts, aiding in the elucidation of complex biological processes and interactions.
Note: For further details, please refer to our recently published paper: Kumar T, Sethuraman R, Mitra S, Ravindran B, Narayanan M. MultiCens: Multilayer network centrality measures to uncover molecular mediators of tissue-tissue communication. PLoS Comput Biol. 2023 Apr 24;19(4):e1011022. doi: 10.1371/journal.pcbi.1011022. PMID: 37093889; PMCID: PMC10159362.
What is centrality measure?
A centrality measure quantifies the importance of nodes within a network, originating from social network analysis and now pivotal in network science across various disciplines. Notable examples include PageRank, which ranks web pages based on their relative importance using the hyperlink structure of the World Wide Web. Centrality measures initially devised for single networks have found broad applications in social, technological, and biological networks. In multilayer networks, these measures can be expanded in diverse ways. Ignoring multilayer structures can yield different ranking outcomes compared to those obtained for multilayer networks, underscoring the significance of considering such structures. To address this, we model multi-tissue data as a multilayer network and propose a tailored set of centrality measures (local, global, and query set) suitable for the multilayer context.
What is local centrality?
In this mode, MultiCens assesses how a node, representing a gene, influences other nodes within the same layer, such as a tissue or a specific region. Local centrality quantifies the impact of a gene on its neighboring genes within a given layer.
What is global centrality?
Unlike local centrality, which focuses solely on within-layer connections, global centrality measures a gene's influence across all nodes in the entire network, disregarding their specific layers. This mode provides a broader perspective on a gene's overall impact within the multilayer network.
What is query set centrality?
The query-set centrality mode evaluates how a gene affects a specific set of nodes within any layer of the multilayer network. It quantifies a gene's influence on a predefined group of nodes, allowing for targeted analysis of its impact on a particular subset within the network.
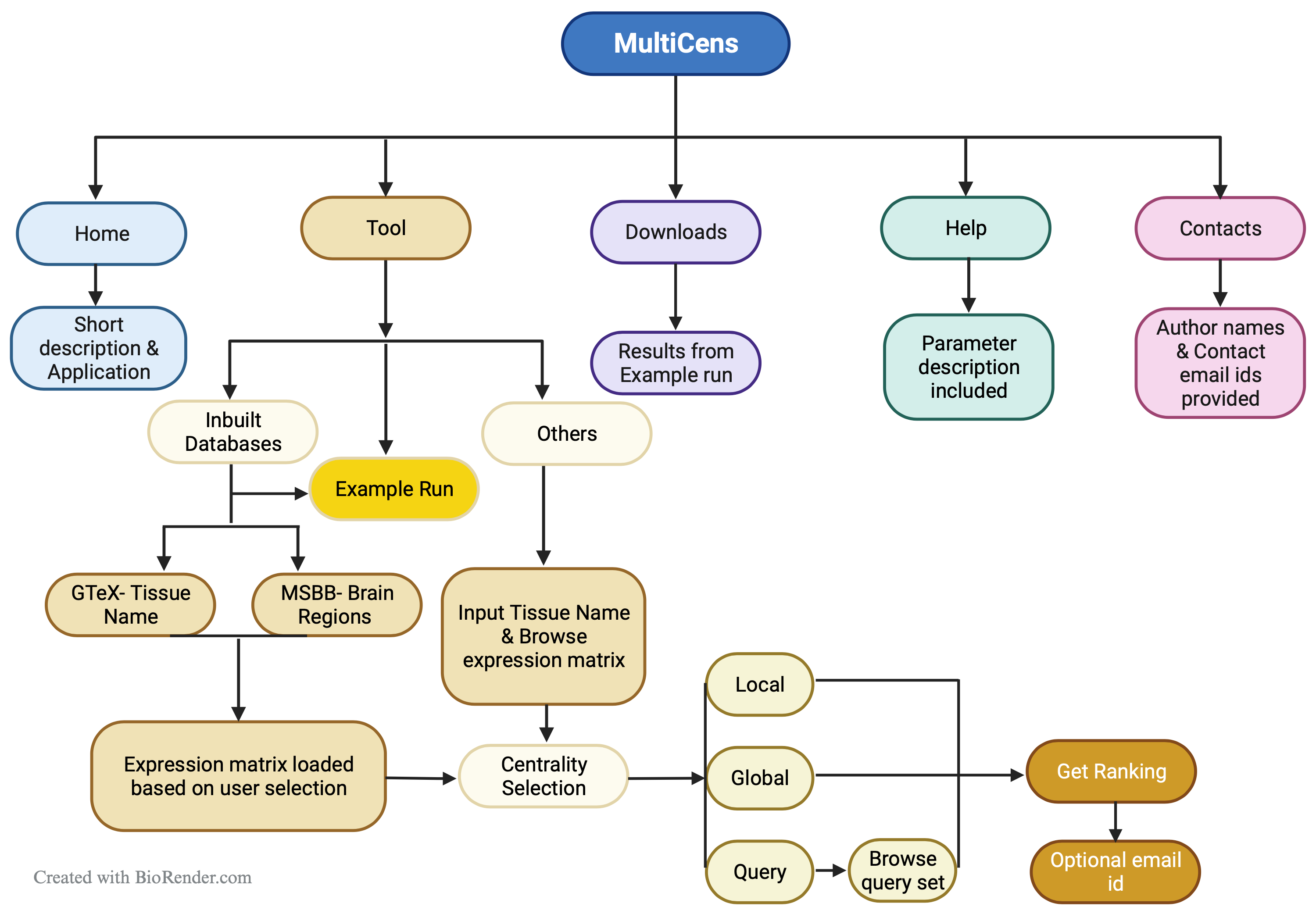
Exploring MultiCens: Example Runs in the Tool Tab
Three examples have been incorporated as three tabs within the "Tool" section, allowing users to click and obtain an overview of how to utilize MultiCens.
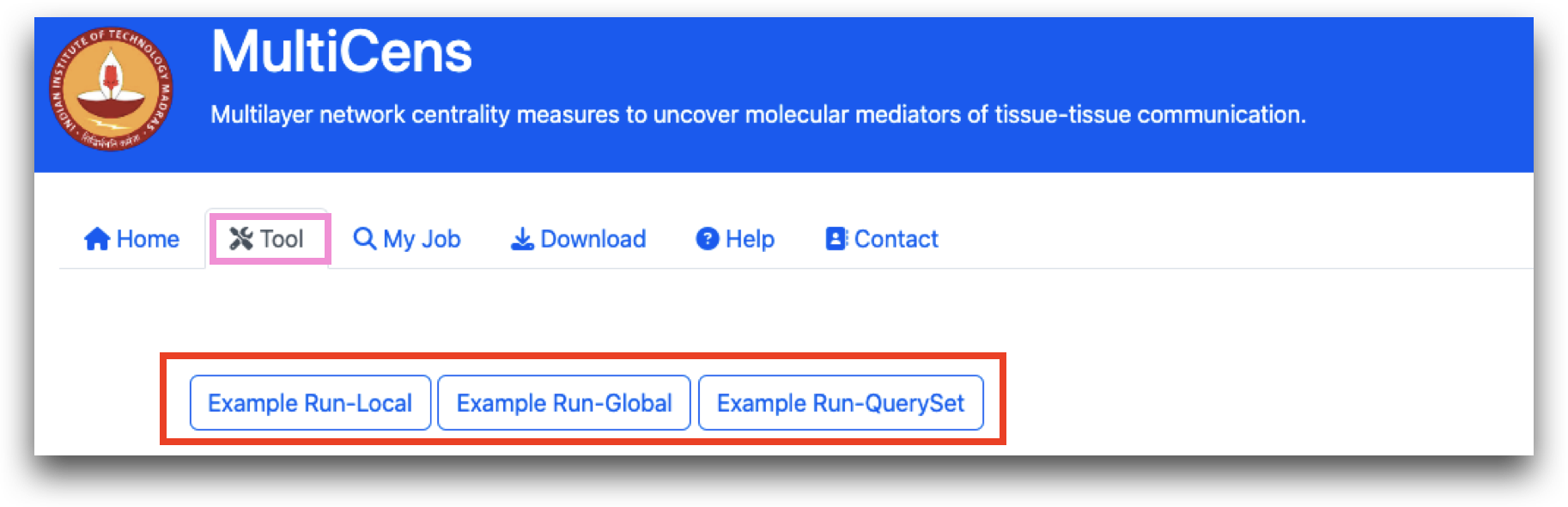
The primary feature of MultiCens is its "Tool" tab. To assess MultiCens' functionality, users must initially choose a dataset and then select tissue names. A minimum of two tissue names is required for testing purposes.
In the MultiCens webserver, gene expression matrices from two known databases (GTEx and MSBB) are already uploaded. Other than that, users can upload their own data.
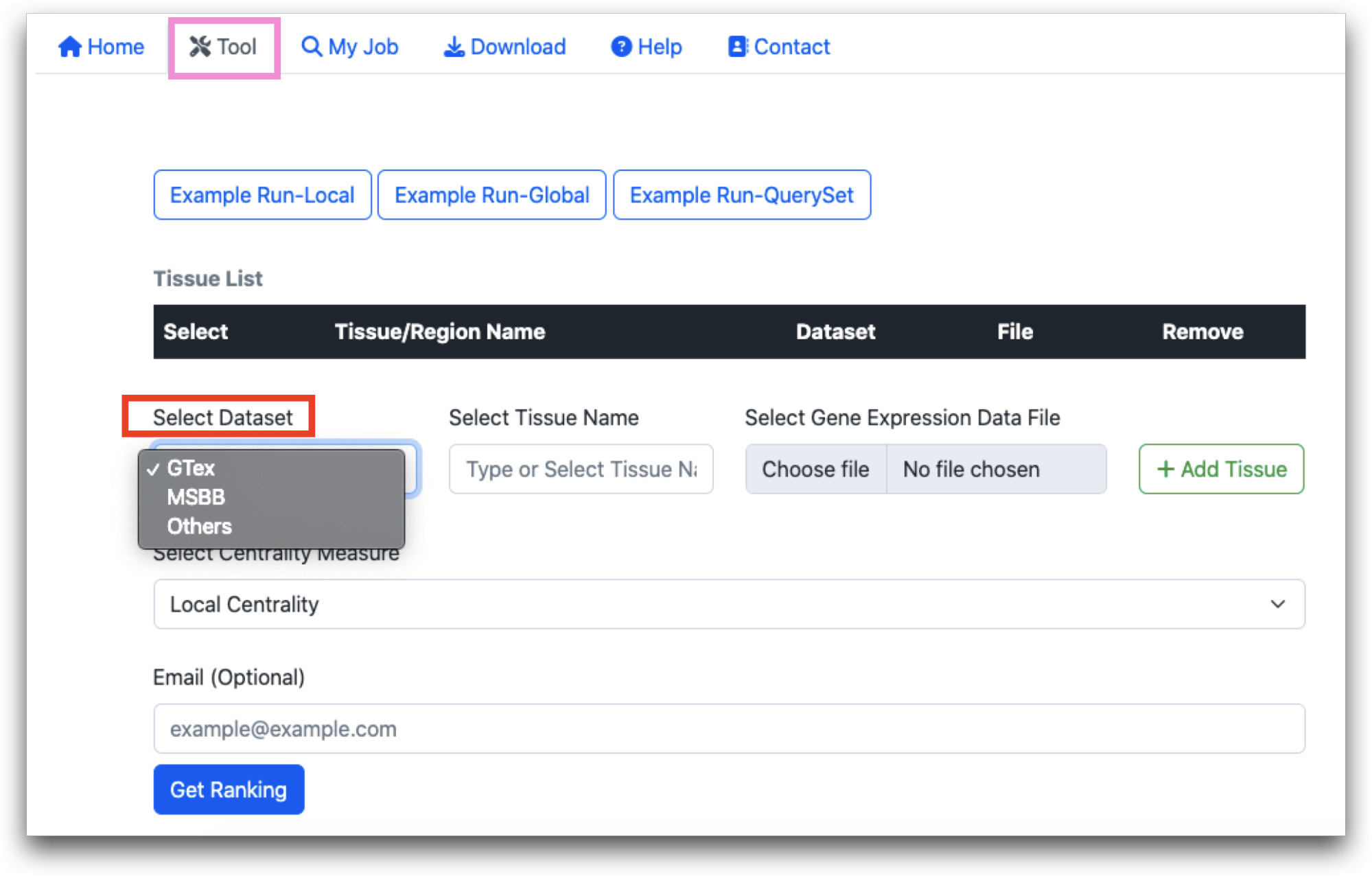
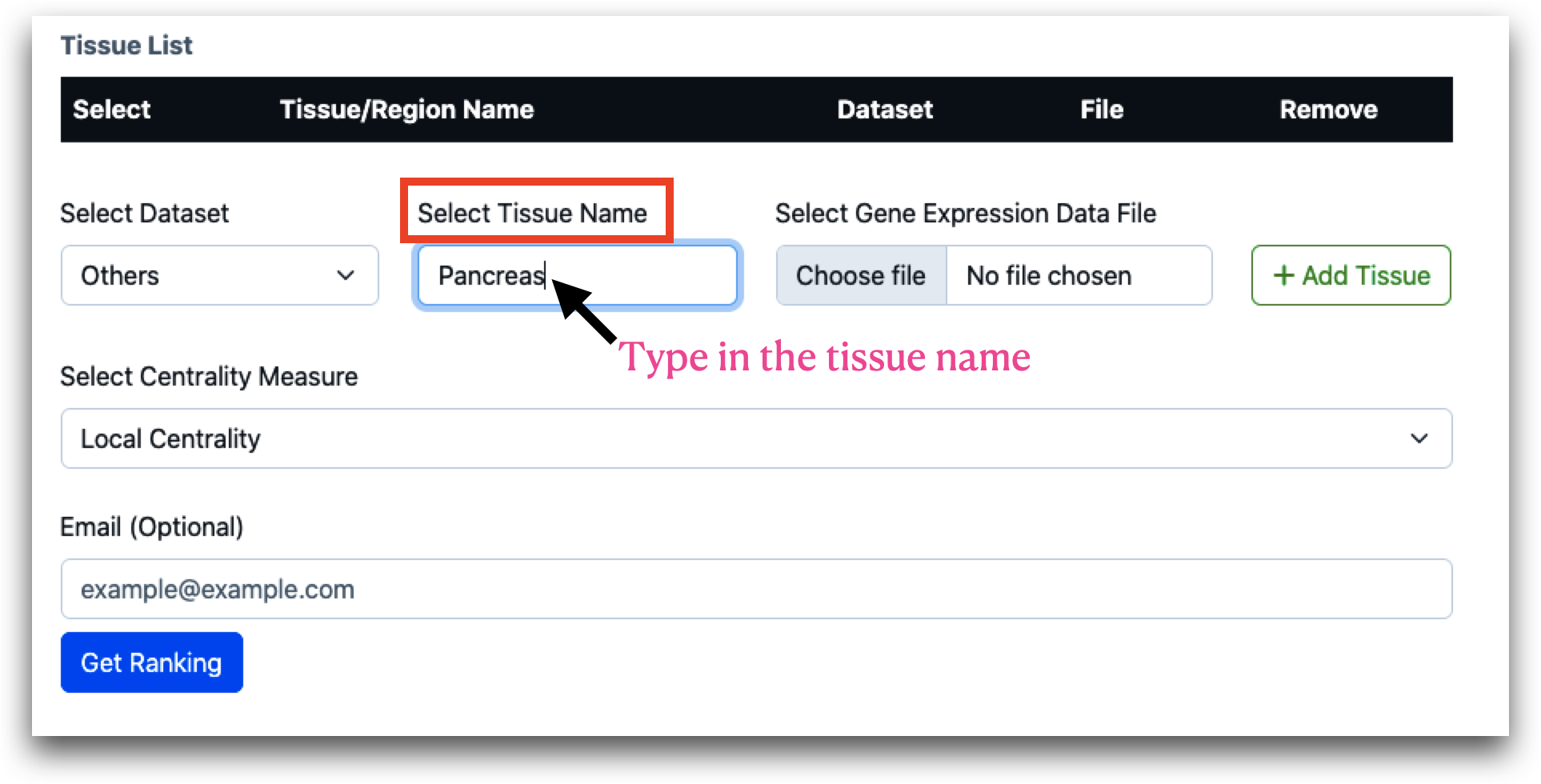
Once the dataset is selected, you must choose the tissue names from the provided list. For GTEx and MSBB, this will be a selection from preloaded options. For other datasets, you will need to manually enter the tissue names.
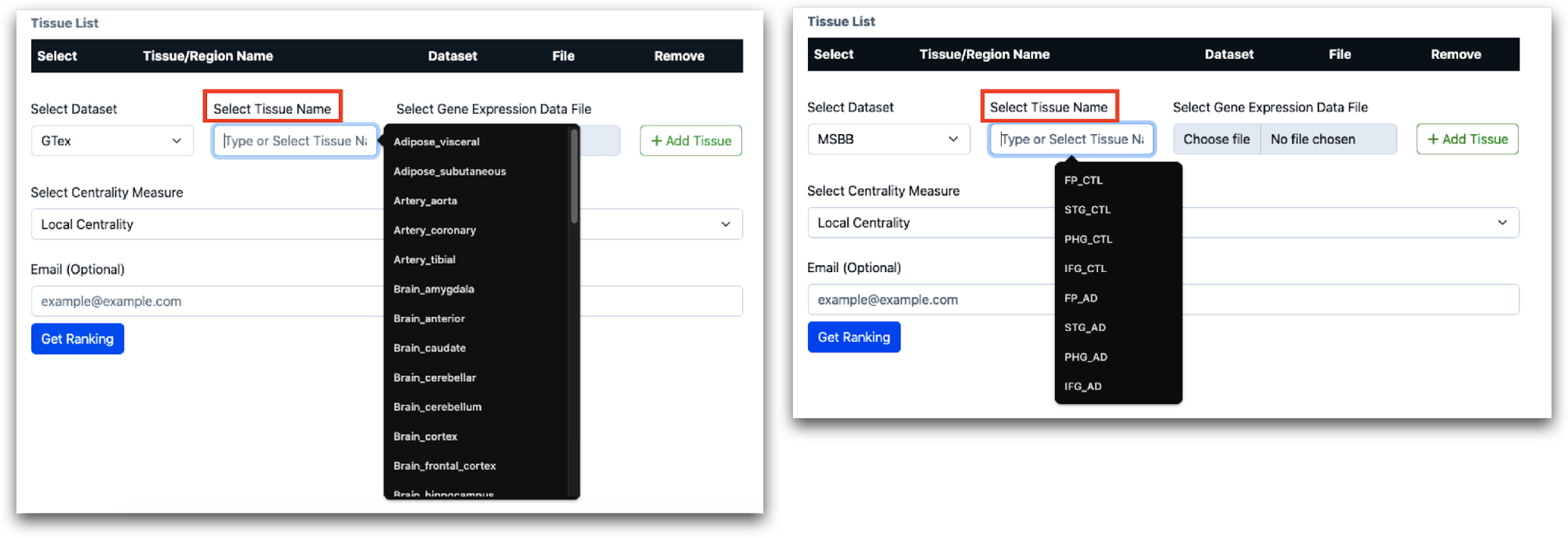
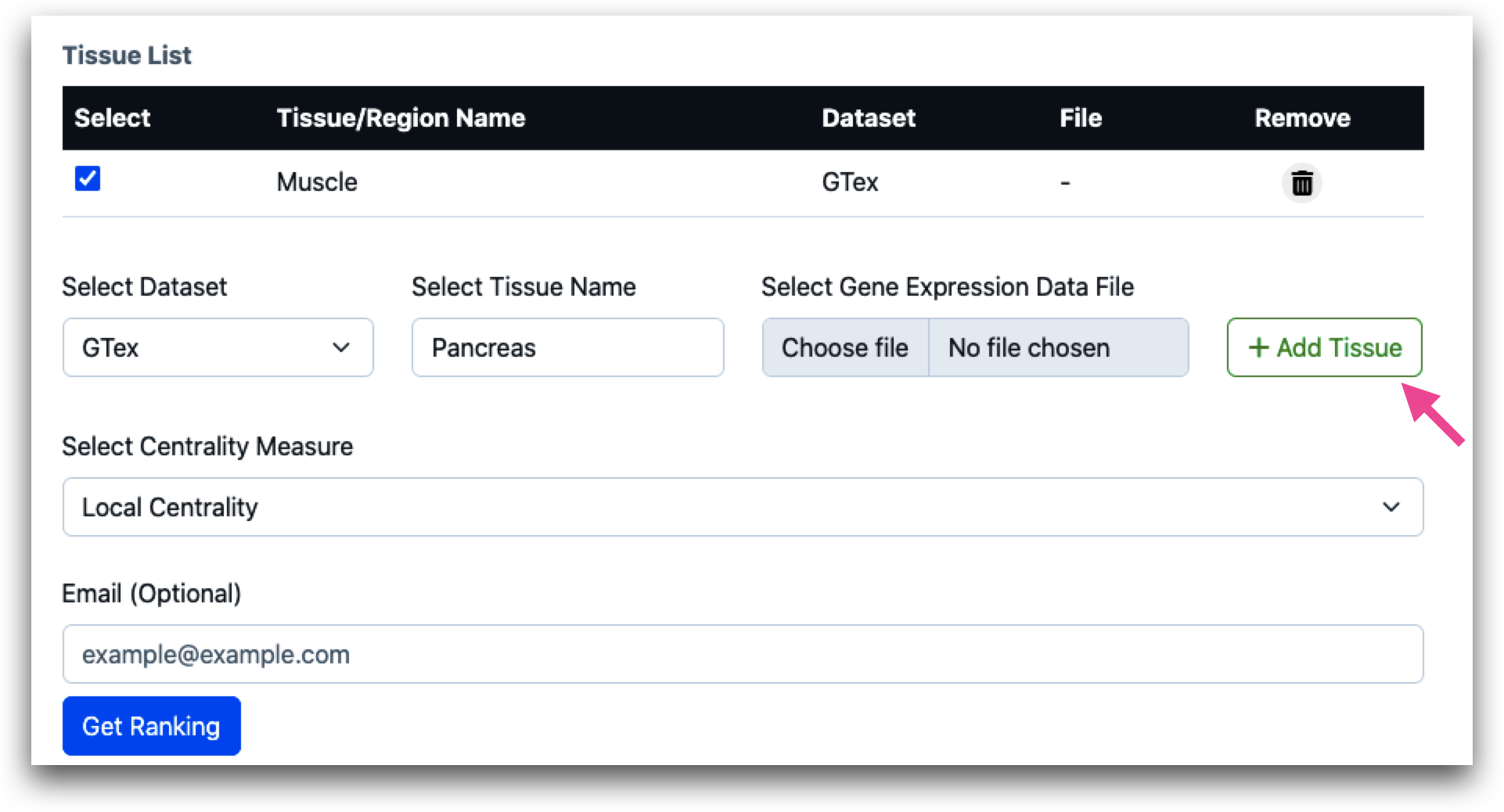
For GTEx and MSBB, since gene expression matrices are preloaded, the “Choose file” option will remain inactive. Users after selecting, tissue name, should directly click “Add Tissue”. The Tissue Name will be added and displayed as shown in the screenshot.
When “Others” is selected under “Dataset”, “Choose file” option becomes active, and users must upload the gene expression data file and click on “Add Tissue”. The data file should be a compressed csv file (.zip or .gz). The maximum file size for each file can be 50 MB.
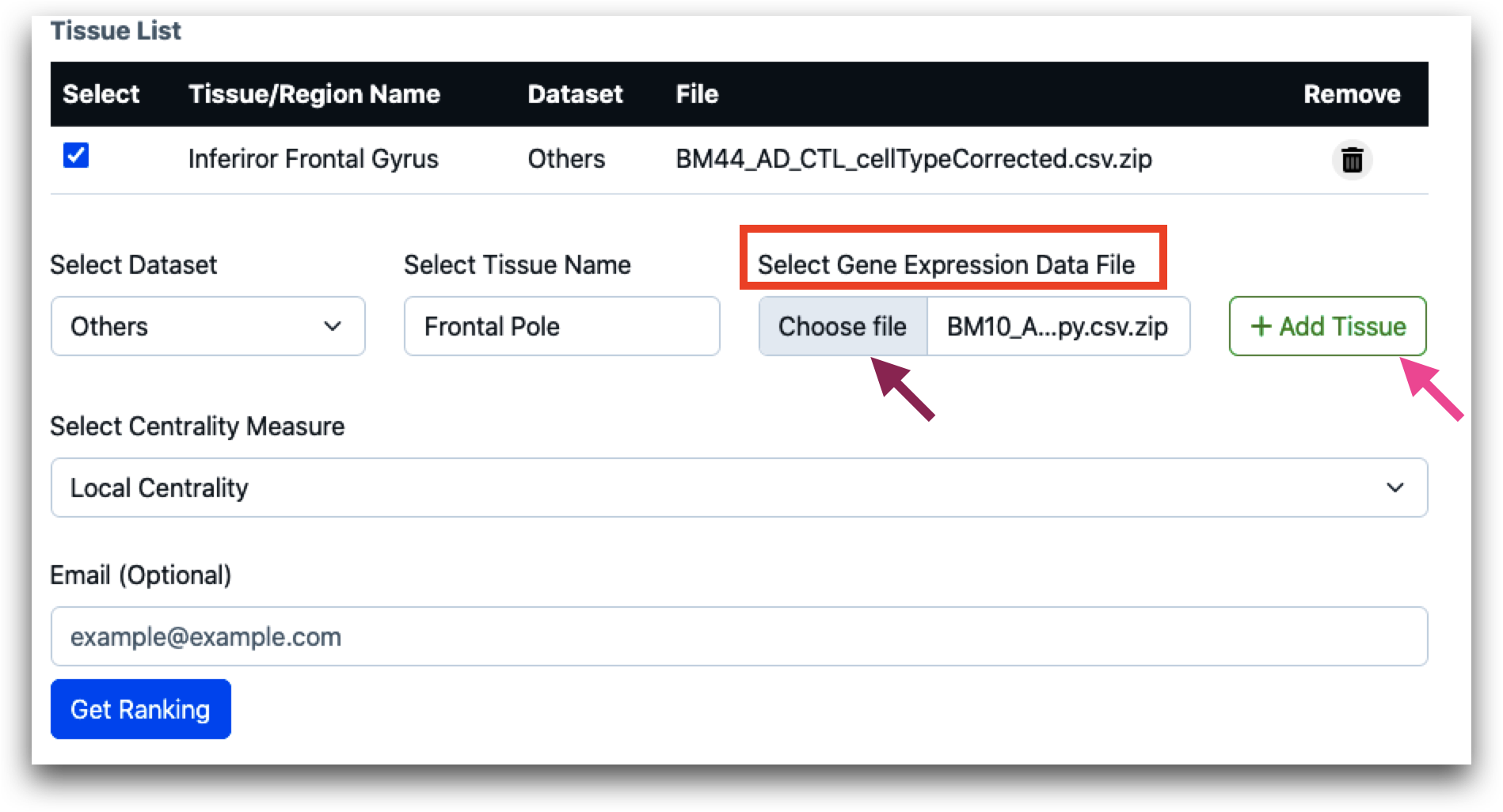
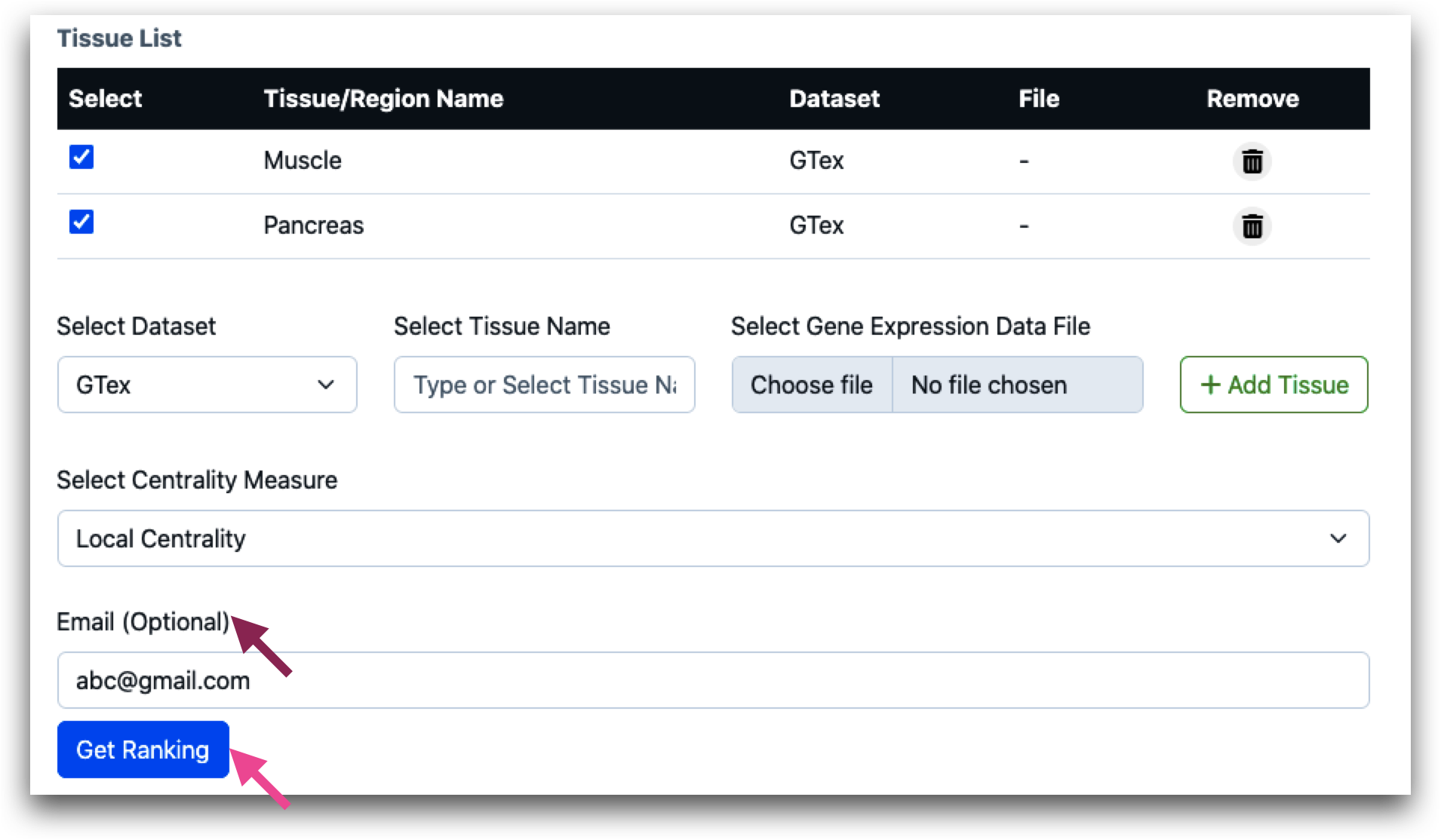
After tissue name is selected, user needs to select the centrality measures. Users can select local, global or query set centrality.
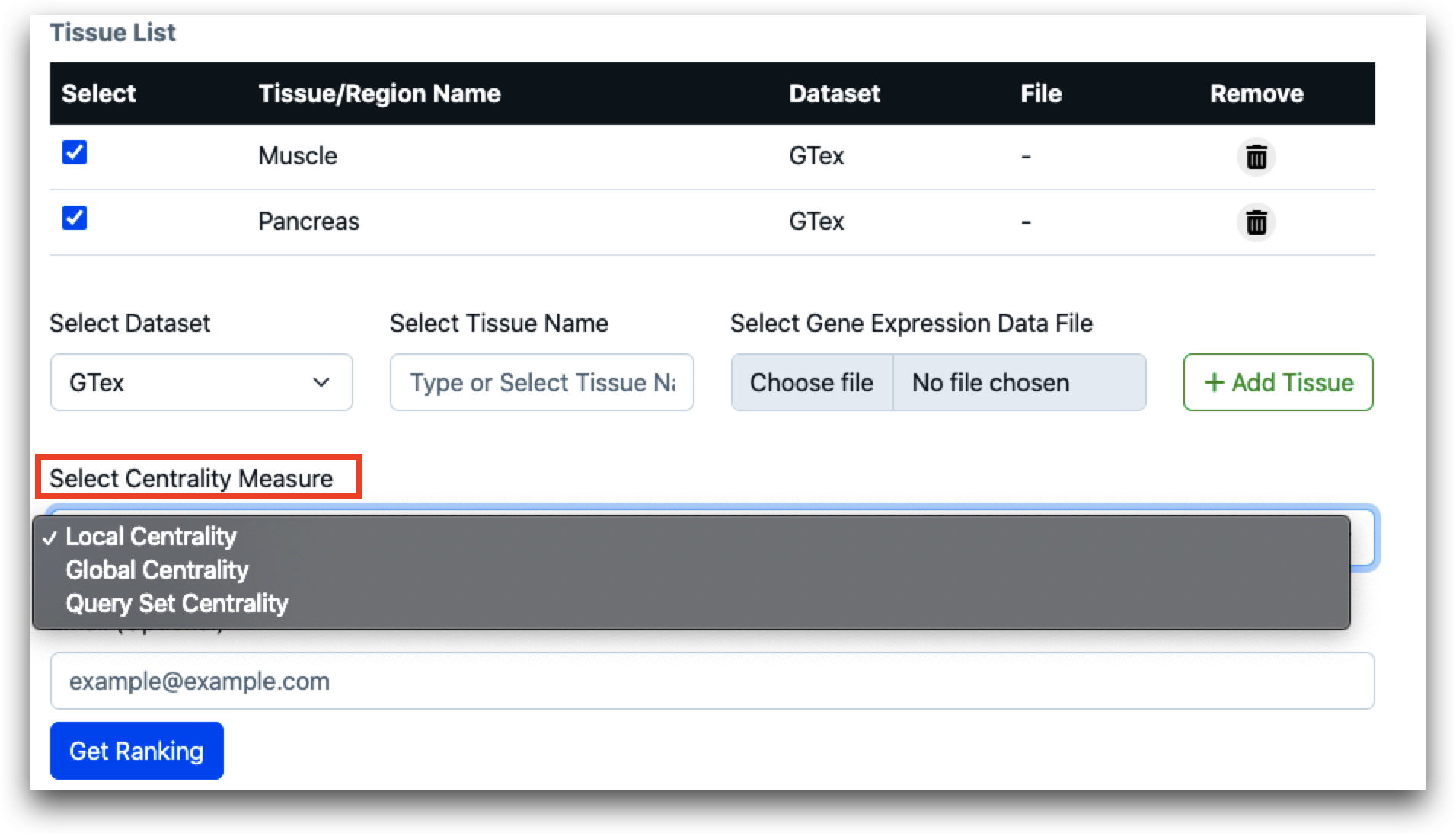
To assess the centrality of a designated gene set (referred to as the query set) associated with a particular biological function or pathway across various layers such as tissues or regions, users can utilize the query set centrality feature. Users are required to upload the query set, which should be formatted in CSV (comma-separated values) format. Additionally, users must specify the tissue name corresponding to the origin of the query set. This enables the tool to accurately analyze and evaluate the centrality of the specified gene set within the specified tissue context.
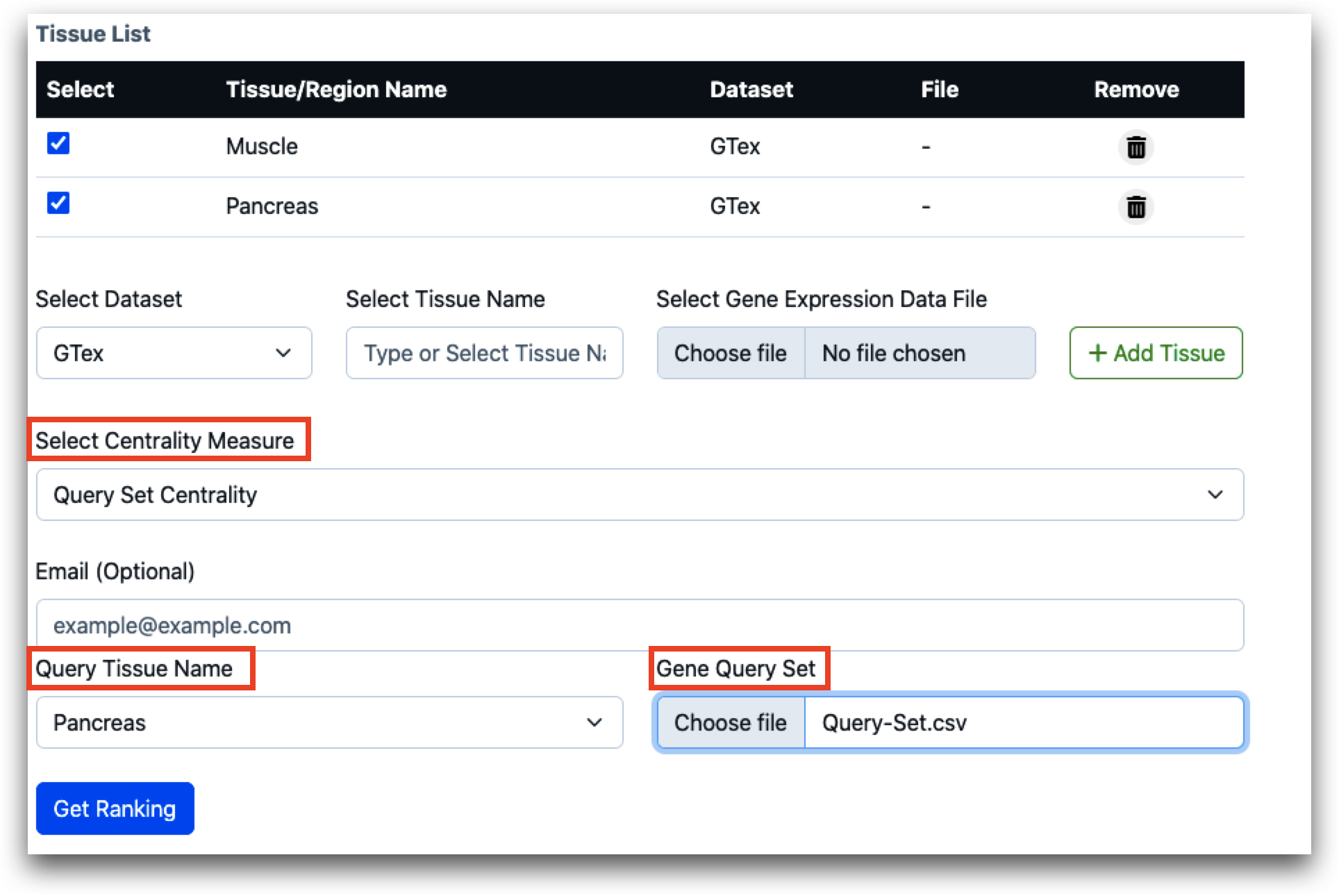
Once all necessary inputs are chosen or uploaded, users simply need to click on the "Get Ranking" button to obtain the result. The tool generates gene rankings according to centrality scores.
In cases where multiple tissues are involved, which might necessitate several minutes to hours for processing, users have the option to provide an email address. This enables the tool to send a URL containing the result to the user for convenient future reference.
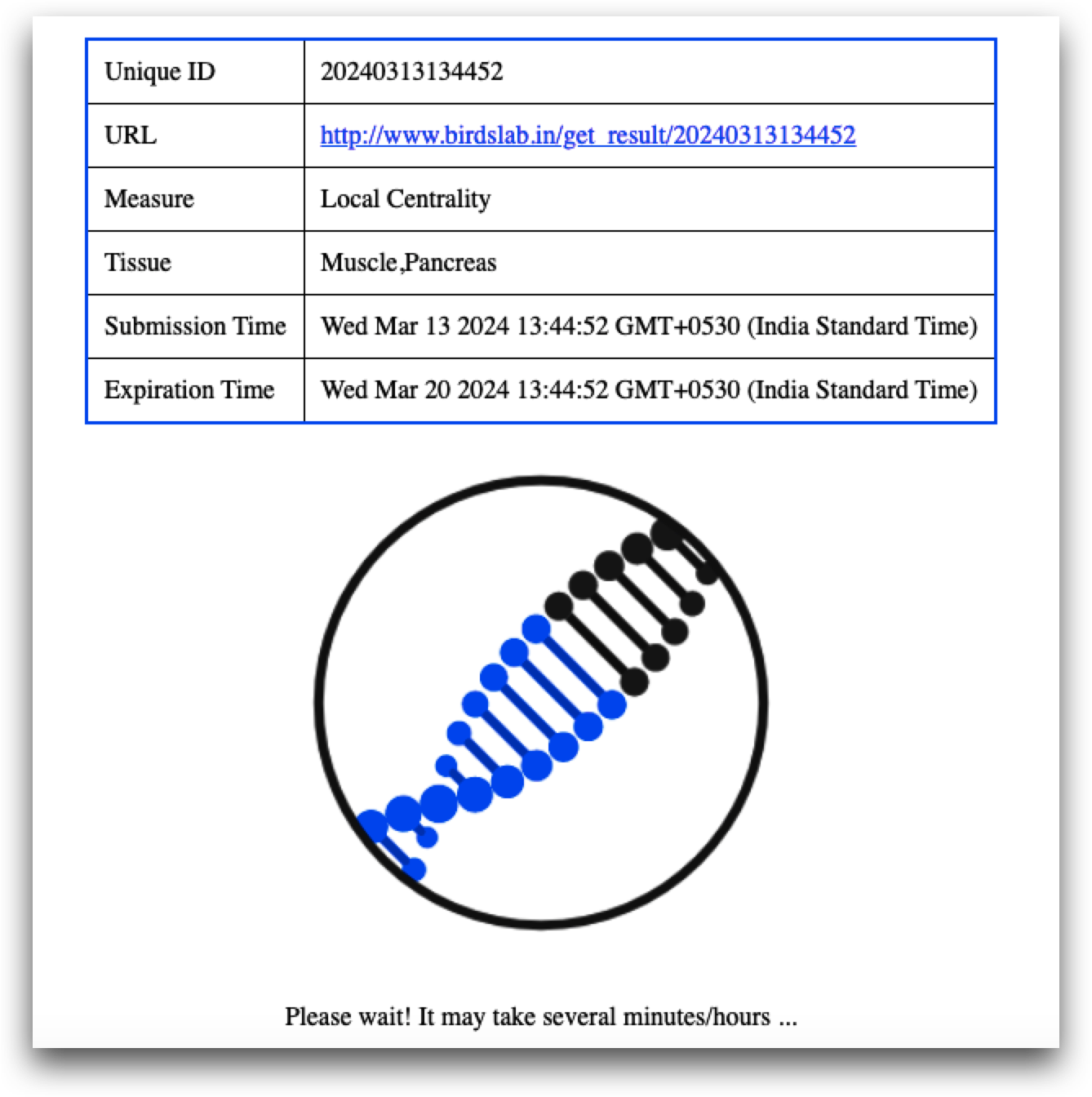
Additionally, a weblink to the result is provided upon data submission, with data stored on the website for seven days. It is imperative for users to download results within this timeframe.
Navigating the MultiCens Result Page
After the result is calculated it will be displayed as follow:
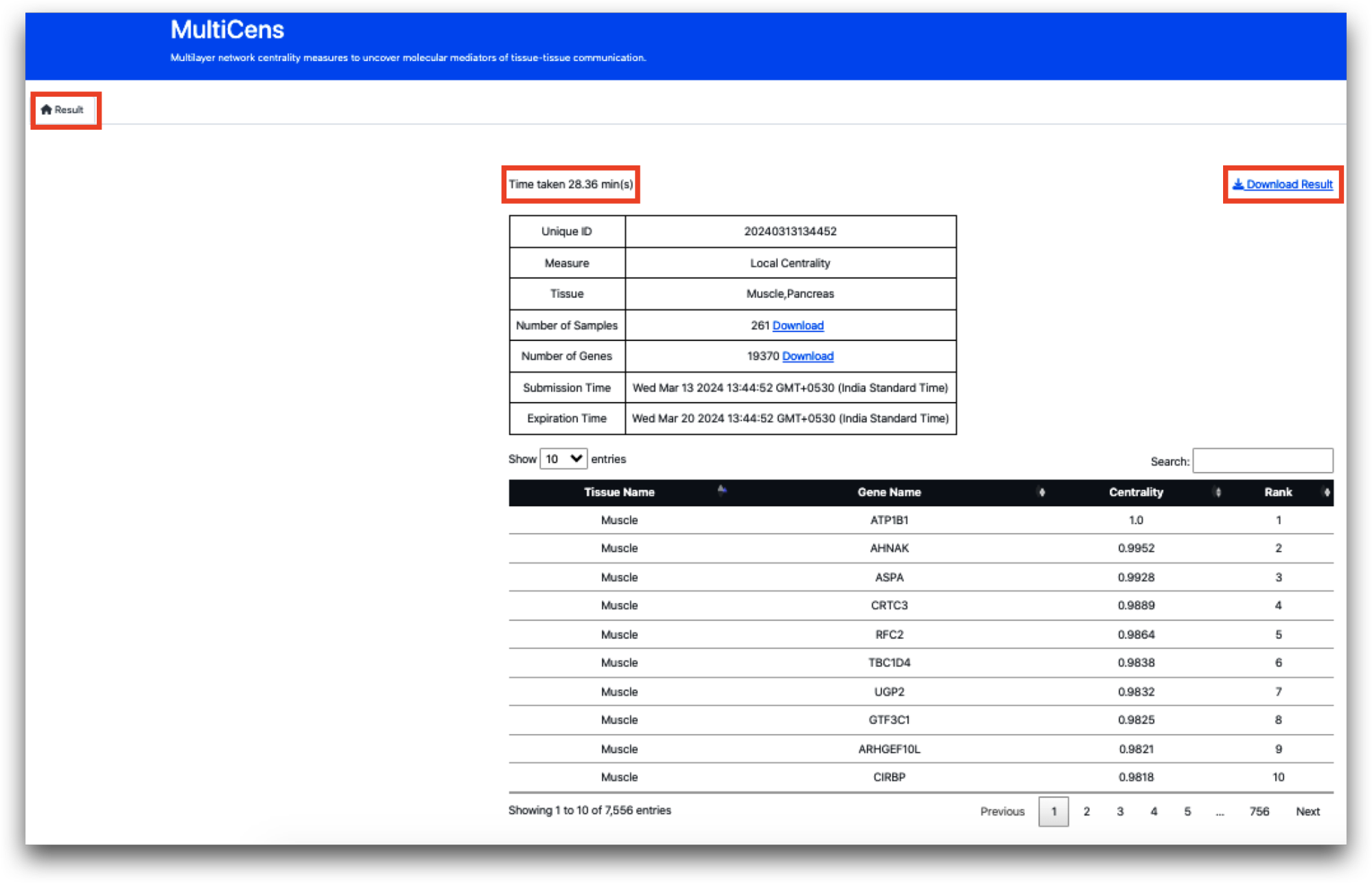
On the Result page, the time taken to complete the task will be prominently shown in the top left corner, while an option to download the result in Excel format will be conveniently placed in the top right corner. Following this, there will be two tables:
The first table records the tissue names and centrality measures provided as input, along with a unique ID for result retrieval within the specified time frame. Additionally, it highlights the number of common samples and genes shared between the given tissues. For instance, between Muscle and Pancreas, 261 common samples and 19,370 common genes are utilized for centrality and gene ranking calculations. Furthermore, the list of common samples and genes can also be downloaded.
The second table, initially displaying 10 entries, showcases the tissue name, gene name, centrality score, and gene ranking. Users have the flexibility to view up to 100 entries at a time, and the table can be sorted based on any of the headers. In total 2000 entries will be displayed on the website. The full list of result containing the top k varying genes by default, will be available for download.
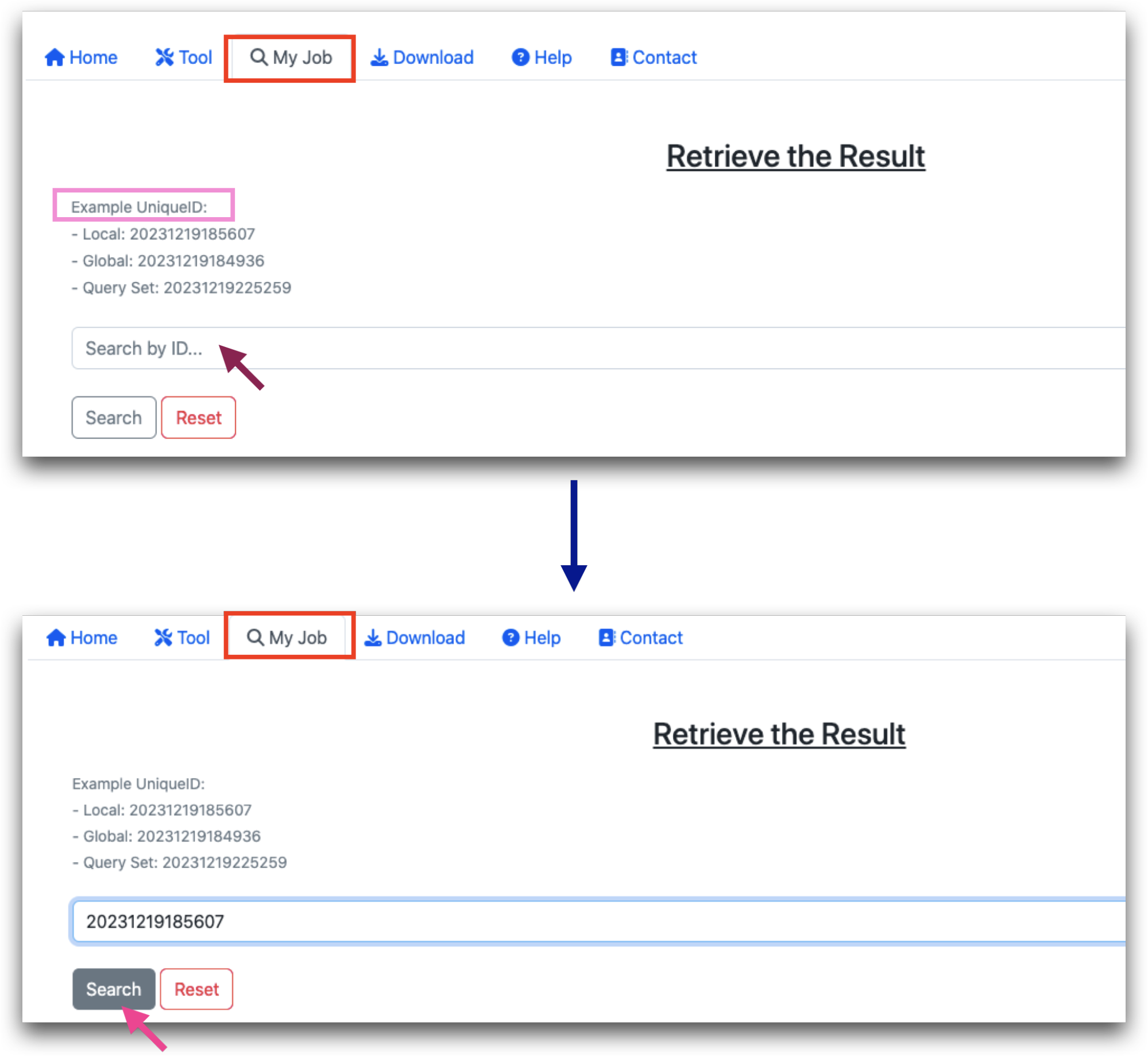
Retrieving MultiCens Result using unique ID
As mentioned above an unique id is generated for each run. This is can be used to retrieve the result for the next seven days from MultiCens website under the “My Job” tab. The id needs to be entered in the search box. A few example ids are also given. The Result page will open in a new tab.
Highlighting the practical application of MultiCens in understanding inter-tissue communication, we utilize the example of tissues involved in insulin production and insulin responsiveness. The pancreas assumes a pivotal role in insulin synthesis and release. Following a meal, when blood glucose levels elevate, specialized beta cells within the pancreas secrete insulin into the bloodstream. Acting as a molecular key, insulin facilitates cellular glucose absorption, thereby reducing blood sugar levels. Insulin acts on various tissues and organs in the body, influencing multiple physiological processes. Among its major targets of action, insulin promotes glucose uptake by muscle cells, facilitating energy production and glycogen storage, while also stimulating the uptake of glucose and promoting fat storage in adipose tissue. In the liver, insulin inhibits glucose production and promotes glycogen synthesis, thereby lowering blood glucose levels. Additionally, insulin plays a role in the brain, where it may regulate appetite and satiety, and in the heart, facilitating glucose uptake to support cardiac muscle function. Furthermore, insulin enhances glucose reabsorption by the kidneys and regulates blood flow in endothelial cells, contributing to overall cardiovascular health. These coordinated effects of insulin on various tissues and organs are essential for maintaining glucose metabolism, lipid metabolism, and overall energy balance in the body, highlighting its critical role in metabolic regulation.
As an illustration, we have examined the gene-gene network within the pancreas and muscle (local centrality), the inter-tissue relationship linking muscle and pancreas (global centrality), and the impact of insulin-related genes moving from pancreas to muscle (query set centrality). The connection between the pancreas and muscle (skeletal muscle) concerning insulin involves the synchronized regulation of insulin secretion by the pancreas and insulin responsiveness in muscle tissue, collectively maintaining glucose homeostasis in the body. Any disruption in pancreatic insulin secretion or muscle insulin response can disrupt this balance, leading to fluctuations in blood glucose levels and potentially contributing to conditions like diabetes mellitus.
First, we present the result table derived from the local centrality analysis. Additionally, we showcase a network visualization created using Cytoscape, delineating the top 25 genes based on rank, in each tissue, namely Muscle and Pancreas. This highlights how the resulting table can be further used for analysis.
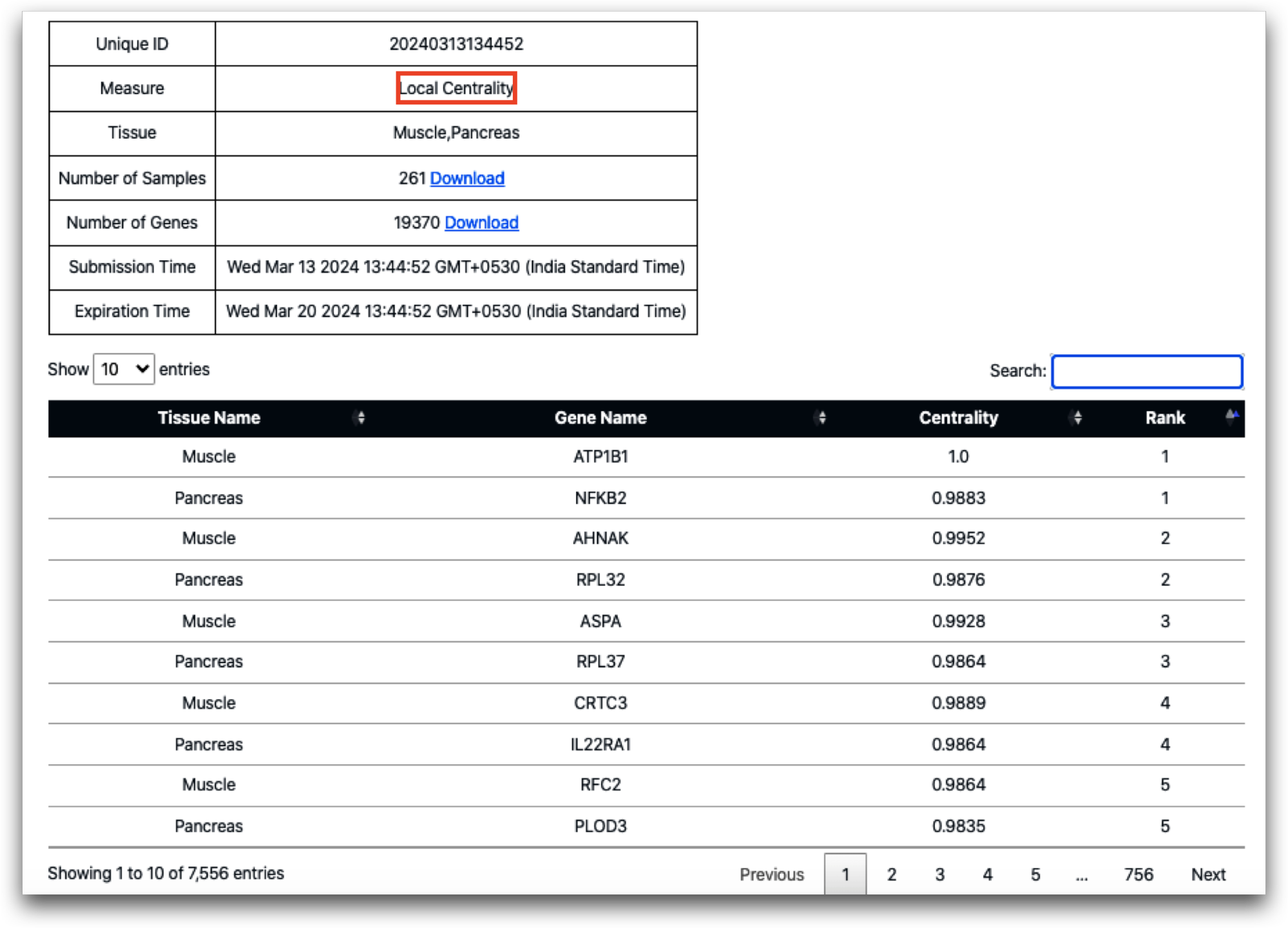
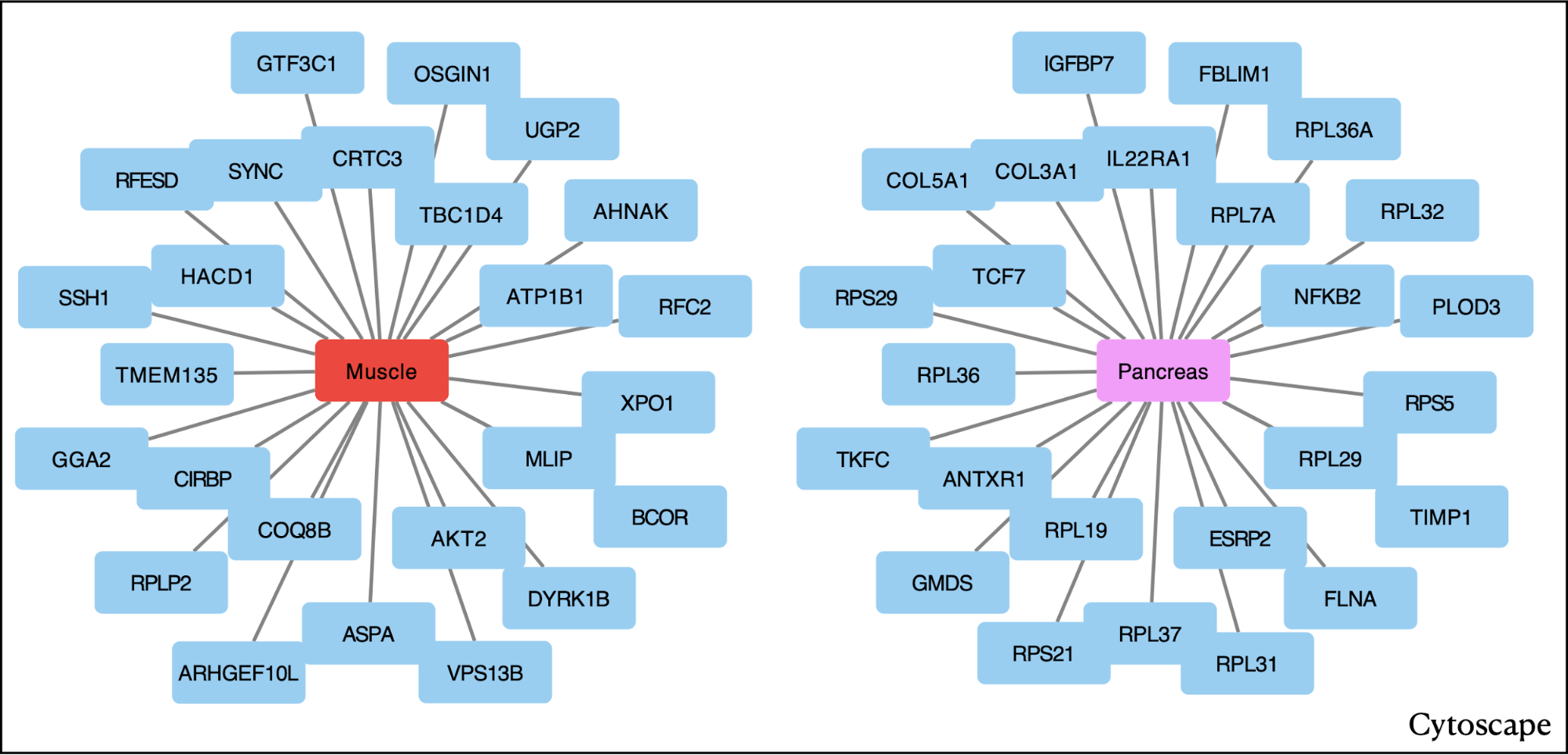
In terms of local centrality measure calculation, the ATP1B1 gene (encoding the ATPase Na+/K+ transporting subunit beta 1) attains the highest centrality score, securing the top rank in muscle. Similarly, the gene NFKB2 (nuclear factor kappa B subunit 2) achieves the highest rank in the pancreas. Notably, in the pancreas, the ATP1B1 gene is ranked 155, while NFKB2 is ranked 2087 in muscle. However, when global centrality is calculated between muscle and pancreas, the centrality measure and rank of these two genes change. ATP1B1 remains among the top-ranked genes in muscle (rank 2), while NFKB2 in the pancreas is ranked 299.
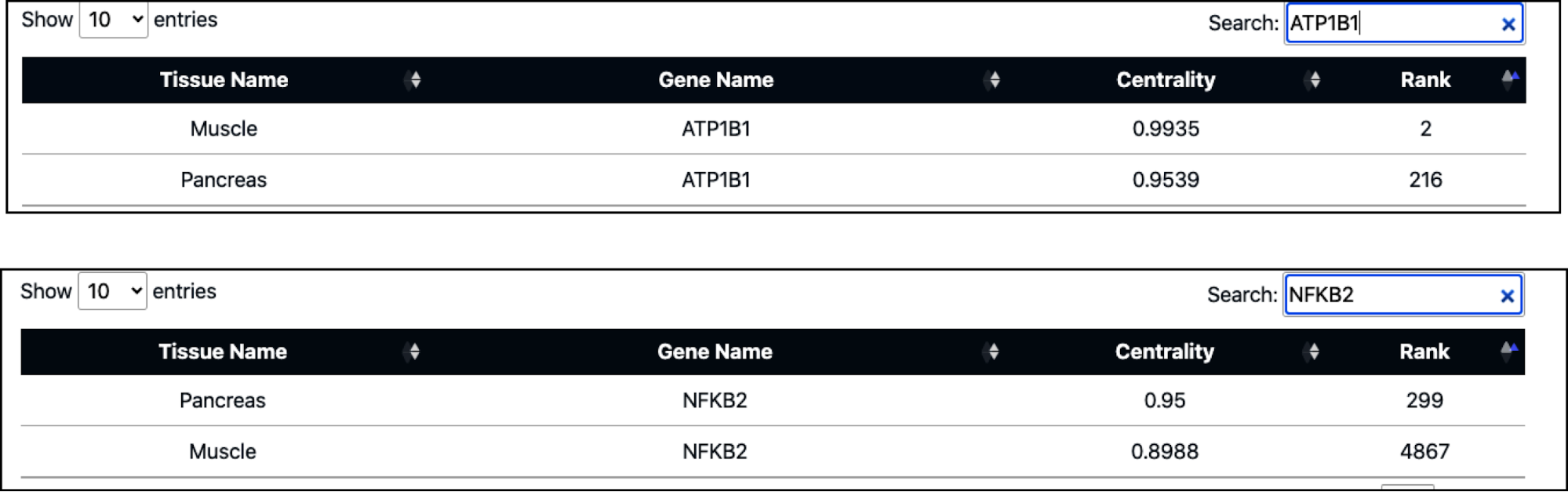
The following list presents the top 10 ranked genes based on global centrality calculation:
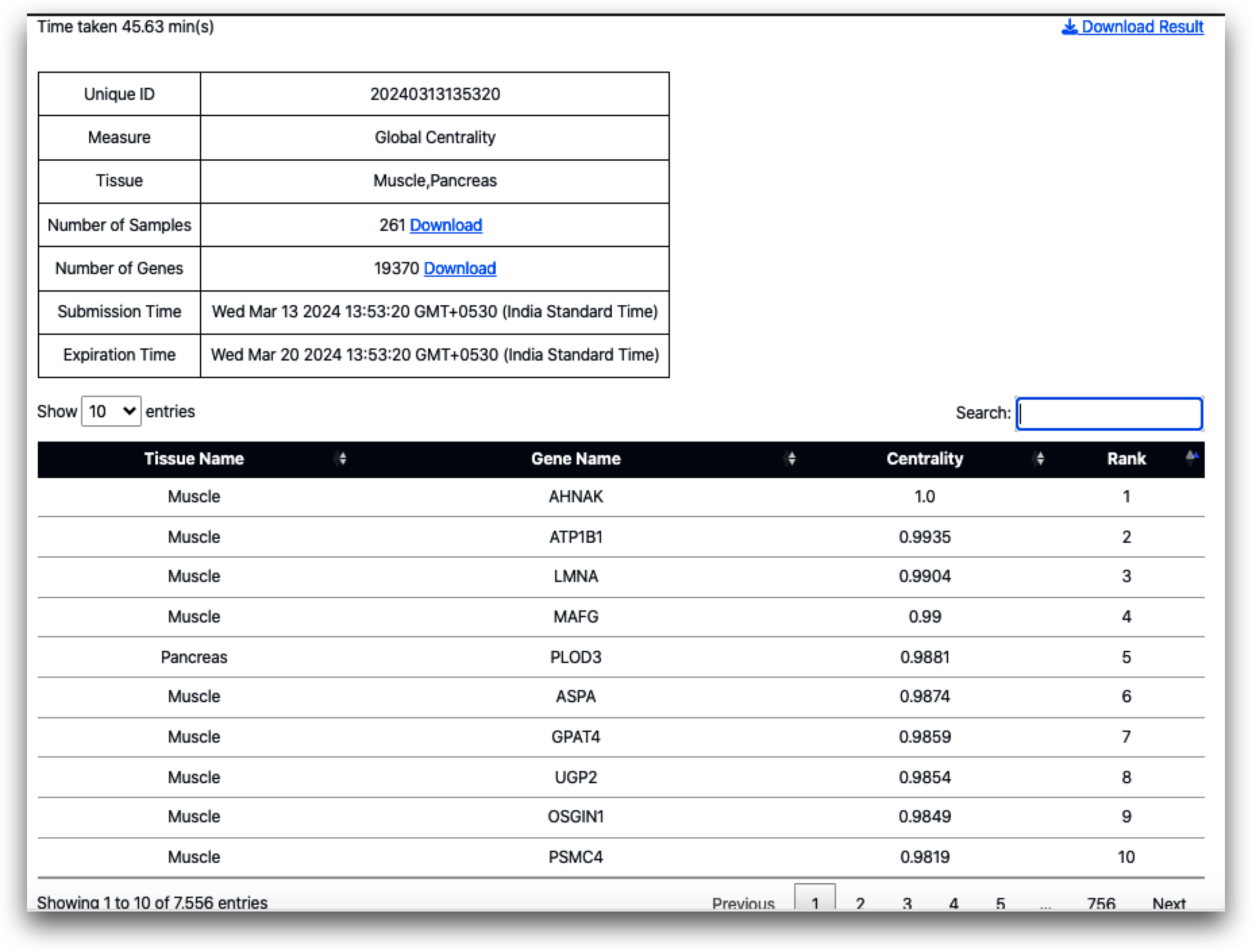
It's intriguing to observe that the AHNAK gene, responsible for encoding the AHNAK nucleoprotein, holds the first rank in global centrality and the second rank in local centrality for muscle. This suggests a significant functional role for this gene in muscle physiology. Notably, among the top 10 genes in global centrality, only one gene, PLOD3 (ranked 5), originates from the pancreas, with the remainder originating from muscle tissue. Interestingly, PLOD3 (procollagen-lysine,2-oxoglutarate 5-dioxygenase 3) also maintains its fifth rank in the pancreas based on local centrality measurement.
Additionally, upon calculating query set centrality, the resulting outcome is as follows (displaying the top 10):
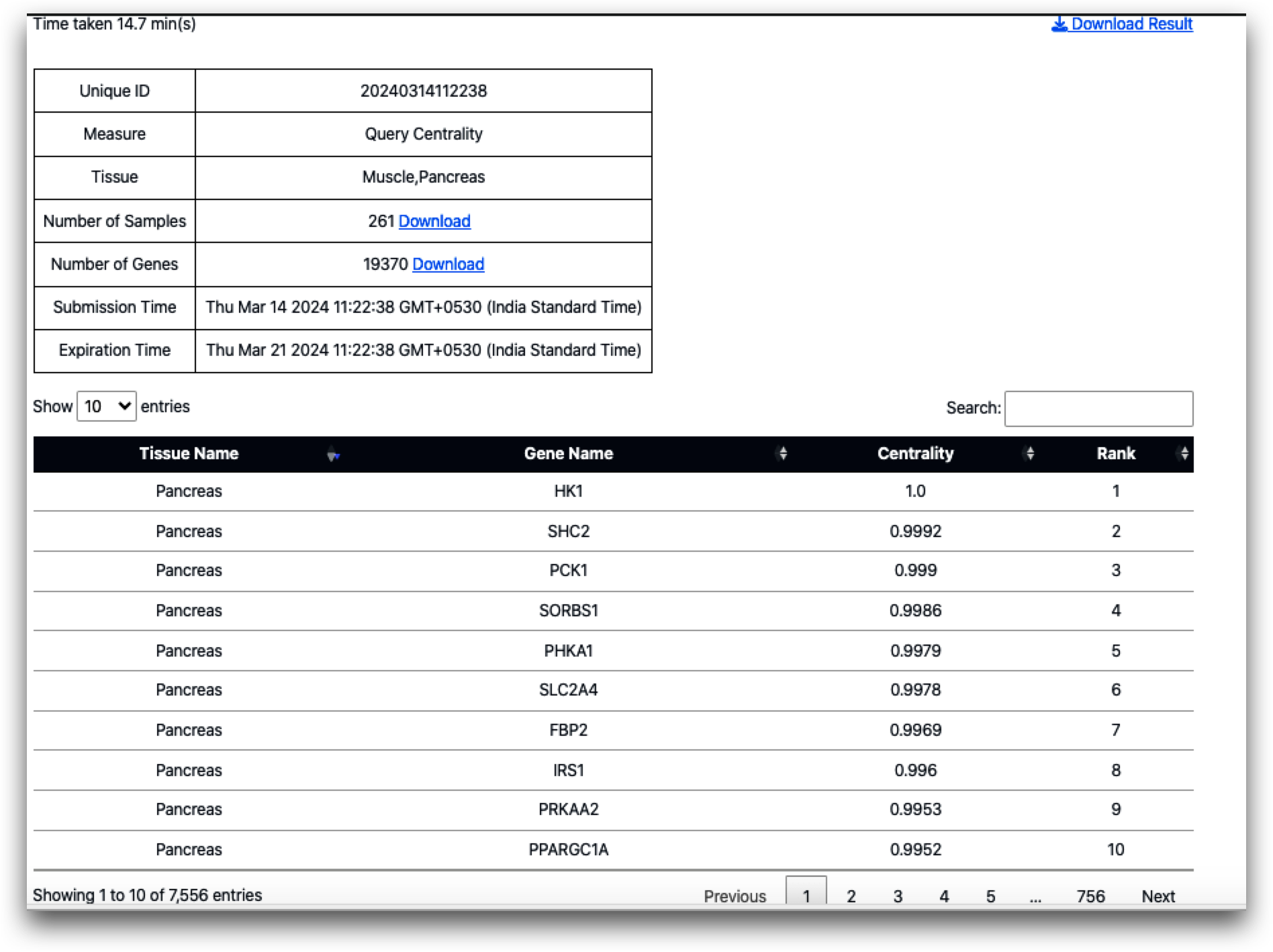
As indicated by the findings, all of the top 10 genes originate from the pancreas, which aligns with expectations given the placement of the query set in the pancreas. Moreover, none of the top 10 genes identified in the query set centrality measures coincide with those identified in the top 10 genes for local or global centrality measures. This observation underscores how inter-tissue communication, influenced by gene-gene networks, can vary depending on the physiological context.
Additionally, it is worth noting that for the same set of tissues, the time required to calculate the three centrality measures varies, with query set centrality requiring the least time and global centrality taking the longest. This relies on the complexities of the underlying multilayer network to calculate centrality scores. The timing also depends on the number of run queued in the system. Based on the load, the same set of tissues for same centrality measure may require different time to execute in different runs.
Watch video tutorials for a step-by-step visual guide through the features and functionalities of MultiCens.
Watch TutorialsThe gene expression matrix uploaded by users should be a compressed CSV file (.zip or .gz). For query set centrality, users are required to upload the query set, which should be formatted in CSV (comma-separated values) format.
It depends on the number of tissues and the centrality measure selected. For two tissues approximately MultiCens takes from 10 - 40 mins. However, email option is provided, so that users can later on retrieve the results either using the URL provided in the email. Further “My Job” tab can be used to access the result using unique ID, given for each submission.
The data is stored for seven days. It can be accessed either using the link provided upon submission or the unique id.
| OS/Browser | Chrome | FireFox | Edge | Safari |
|---|---|---|---|---|
| Windows | Yes | Yes | Yes | No |
| MacOS | Yes | Yes | No | Yes |
| Linux | Yes | Yes | No | No |
Yes. The complete MultiCens source code is available on GitHub: https://github.com/KartikGondalia/Multicens
At present, we have only tested on humans. In the future, we intend to add data for other species.
MultiCens is open for all users, both academic and non-academic free of charge. There is no login requirement. We appreciate if you share your excitement about our service with your colleagues and cite us when you have found our service useful.
This resource is intended purely for research purposes. It should not be used for emergencies or medical or professional advice.
The Multicens Centrality Analysis API provides computational analysis of gene expression data to determine various centrality measures across different tissues. This service is essential for researchers and biologists aiming to identify central or key genes within a tissue dataset.
Base URL: https://www.birdslab.in
Authentication: The API currently does not require authentication.
HTTP Methods:
This endpoint allows for the submission of gene expression data files along with metadata to compute local, global, or query-set centrality measures.
| Parameter | Type | Required | Description |
|---|---|---|---|
| measure | string | Yes | The type of centrality measure to compute. Accepted values: local, global, query. |
| string | No | Optional. Email address for receiving job status notifications and links to the results. | |
| dataset | string | Yes | Specifies the dataset used for analysis. Accepted values: GTEx, MSBB, or custom. |
| tissue | string | Yes | Specifies the tissue name involved in the analysis. Must be compatible with the selected dataset. |
| files | file | Yes | Gene expression data files in .zip or .gz format. |
| Parameter | Type | Required | Description |
|---|---|---|---|
| query_tissue | string | Yes | Specifies the tissue name from the previously selected tissues for performing the query analysis. |
| query_gene_file | file | Yes | A CSV file containing the list of genes to include in the query centrality calculation. |
curl -X POST "https://www.birdslab.in/get_centrality" \
-H "Content-Type: multipart/form-data" \
-F "measure=local" \
-F "email=example@example.com" \
-F "dataset=GTEx" \
-F "tissue=heart" \
-F "files=@heart_data.zip"{
"message": "Result will be displayed. Check your email for results link or use your uniqueID to track the status.",
"uniqueID": "12345ABC",
"statusUrl": "https://www.birdslab.in/get_result/20240410131556"
}{
"error": "Unable to process request due to missing files.",
"statusCode": 400
}This endpoint retrieves the results of a centrality analysis job using a unique identifier provided at the time of job submission. The results include detailed centrality rankings for genes within specified tissues, if the analysis has been completed within the allowed time frame.
GET https://www.birdslab.in/get_result/20240410131556{
"status": "Complete",
"results": "Link to download the analysis results."
}404 Not Found: "No result found for this uniqueID"500 Internal Server Error: "Database error"400 Bad Request: "Your result calculation has exceeded the maximum time limit"This endpoint facilitates the downloading of pre-computed centrality results. It provides users with access to stored results based on various centrality measures computed on standard datasets such as GTEx.
This endpoint ensures that users can securely download pre-computed result files, which include detailed analyses of different tissues or combinations of tissues under various centrality measures.
404 Not Found: If the filename does not exist or is not available.400 Bad Request: If the filename is not secure or has been tampered with.GET https://www.birdslab.in/download/example_results.csvThe download functionality is accessed through the ‘/download’ tab on the main site. A table lists all available pre-computed result files. Each row provides details about the tissue/region name, centrality measure, dataset used, and a direct download link.
File Links are managed through url_for('download_file', filename='filename.csv'), where ‘filename’ is the name of the result file.
@app.route('/download/<filename>', methods=['GET'])
def download_file(filename):
filename = secure_filename(filename)
filepath = os.path.join(app.root_path, 'static/sampleData', filename)
try:
return send_file(filepath, as_attachment=True)
except FileNotFoundError:
return "File not found", 404
except Exception as e:
return str(e), 500
The API is rate-limited to 5 requests per hour per IP to prevent abuse and ensure availability. Exceeding this limit results in a ‘429 Too Many Requests’ status code.
For further assistance or to report issues, please contact birdslab.iitm@gmail.com. Our team is committed to ensuring a smooth and secure user experience.
Associate Professor,
Dept. of Computer Science & Engg,
Indian Institute of Technology Madras,
Email: nmanik@cse.iitm.ac.in
Intel PhD fellow,
Dept. of Computer Science & Engg,
Indian Institute of Technology Madras,
Email: cs15d017@cse.iitm.ac.in
Senior Research Scientist,
Dept. of Computer Science & Engg,
Indian Institute of Technology Madras,
Email: 011141@imail.iitm.ac.in
M.Tech. Student 2024,
Dept. of Computer Science & Engg,
Indian Institute of Technology Madras,
Email: kartikgondaliya0@gmail.com
M.Tech. Student 2023,
Dept. of Computer Science & Engg,
Indian Institute of Technology Madras,
Email: cs20m052@smail.iitm.ac.in